In recent years genome-wide association studies (GWAS) have discovered thousands of genetic variants associated both with diseases and complex traits.
Geneticists had hoped these studies might transform precision medicine by enabling individualized disease risk prediction and treatments tailored to the variations in a person’s genes (a field known as pharmacogenomics). Most common diseases, however, are not caused by single genetic mutations, which are far more accessible to precision medicine strategies. Instead, GWAS have revealed that the majority of common diseases have a polygenic architecture, wherein multiple genetic variants, each by themselves of small effect, cumulatively impact disease risk.
Advances in statistical and clinical genetics have started to demonstrate the power of polygenic risk profiling in the form of polygenic risk scores to identify individuals at higher (and lower) risk of disease. This article reviews recent developments in polygenic risk profiling and the predictive utility of polygenic risk scores to uncover a person’s susceptibility to disease.
The Promise of Genetics and Genomics for Medicine
During the past decade, significant progress in genomic technologies has allowed scientists and clinicians to explore the human genome in incredible detail. Consequently, the growth in our understanding of genetics and genomics may have the potential to transform all aspects of precision medicine, including prevention of disease manifestation, accurate diagnosis and prognosis of disease, pharmacogenomics, and motivating lifestyle changes to improve health.
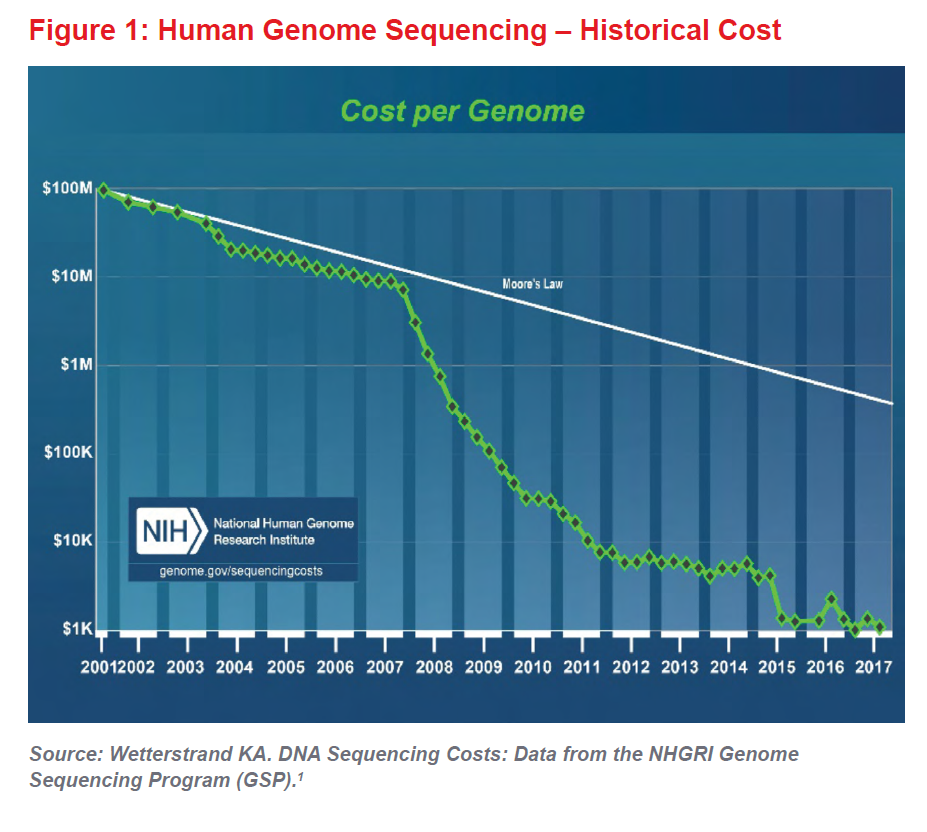
When coupled with the ever-decreasing cost of DNA sequencing (see Figure 1), the era of genomic medicine can now be considered truly under way. The first human genome cost $2.7 billion and took almost 15 years to sequence. Now sequencing costs about $1,000 and can be done in a few days. In a few years, some believe the whole genome will be sequenced for as little as $100. As research continues to advance our understanding of genetics, genomic medicine will almost certainly lead to improvements in mortality and longevity, which will be positive for the life insurance industry and for society as a whole.
What is a Polygenic Risk Score?
A polygenic risk score (PRS) is a metric that condenses information from tens, hundreds, thousands or even millions (“poly”) of a person’s genetic variants (“genic”) into a score that measures the individual’s genetic predisposition to specific diseases or complex traits.
Genome-wide association studies (GWAS) are a relatively new way for geneticists to search among the millions of variants in the human genome and identify those associated with certain diseases or complex traits. These genetic variants are known as single nucleotide polymorphisms or SNPs (pronounced “snips”). DNA has four different base nucleotides (DNA building blocks) and a SNP occurs when a nucleotide in a particular position in the genome is different
Geneticists have observed that certain SNPs occur more commonly in people with a particular disease. Over the 13 years since the first GWAS was conducted (2005),2 millions of SNPs have been found and isolated. These SNPs have been shown to increase or decrease a person’s risk of developing complex diseases such as diabetes,3 breast cancer,4 and cardiovascular disease.5
Clinicians commonly test for high-penetrance single gene mutations such as BRCA1 or BRCA2, two genes that confer increased risk for breast cancer. Such simple one-to-one relationships between a gene and susceptibility to disease, however, are very rare. Rather, the genetic architecture of most disease is overwhelmingly polygenic, with multiple SNPs, each of small effect, cumulatively affecting disease risk. No single SNP can be used in prediction, as the increase or decrease in risk is extremely small. However, merging information across thousands or even millions of SNPs can be useful in predicting disease risk.
It is hoped that as GWAS grow in sample size and more SNPs are investigated, researchers will uncover additional SNPs associated with common diseases as well as SNPs that can influence a person’s response to medications.
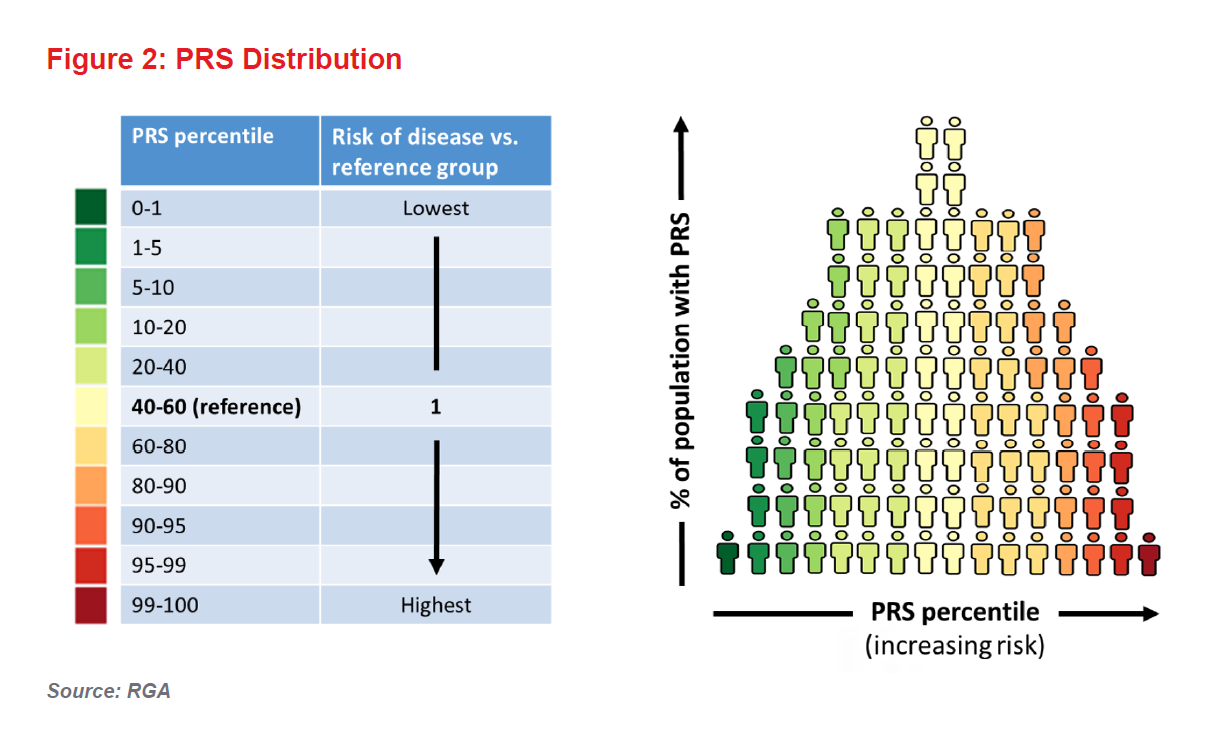
And so we come to PRSs. These scores, which are calculated based on an individual’s SNPs, are generally expressed as risk percentiles correlated against the risk of particular disease in a given population. They are already proving most useful in terms of risk predictions in the extremes of the genetic risk distribution (see Figure 2).
An individual with a PRS in the 100th percentile for diabetes, for example, would be considered to have the highest genetic risk. However, for an individual with a PRS closer to the population mean, the score would offer little additional risk information (i.e., the person’s predicted risk, based on his or her genetics, will be similar to the population’s average risk).
In the past few years, academic researchers have developed PRSs for many conditions and have tested them in their studies. The pertinent question, however, remains: will PRSs transform clinical risk assessment in terms of disease prediction, stratification, and prognosis? And could they impact insurance medicine, underwriting, product development, and claims experience?
Clinical Utility of Polygenic Risk Scores and Possible Applications
As PRSs are simple to calculate and remain constant over a lifetime, their potential in clinical medicine is vast. Several studies suggest PRSs could be integrated into clinical risk tools in the near future. For example, a PRS to predict coronary artery disease risk has already demonstrated that people with a PRS in the highest 5% have a threefold increased risk of experiencing the condition compared to individuals with lower PRSs.6In a study on genetic risk and breast cancer, women whose PRSs were in the top 20% were shown to have a 17.2% lifetime risk of breast cancer compared to a 5.3% lifetime risk for women whose scores were in the lowest quintile.4
Another application of PRSs could be to redefine cancer screening algorithms. For instance, women with PRSs indicating higher breast cancer risk could be offered more regular cancer screenings. In the United Kingdom, breast cancer screening is first initiated in women 47 years of age. This screening decision is based on the 10-year risk of breast cancer in the average woman. Researchers, however, have now illustrated that women with PRSs in the 96th to 100th percentile attain this level of risk at just 37 years of age, while those with PRSs below the 20th percentile will never attain this level of risk in their lifetimes.4
Of course genetics only forms part of a person’s disease risk profile. Environmental factors clearly also contribute, and an additive relationship is generally seen between these. For example, in coronary disease, an individual with a “healthy” lifestyle but at the highest level of genetic risk (i.e., a PRS in the highest quintile) has roughly the same risk of suffering a coronary event as does an “unhealthy” individual with the lowest level of genetic risk (i.e., a PRS in the lowest quintile).6 “Healthy” and “unhealthy” are defined here in terms of the individual’s smoking, diet, and exercise habits and body mass index.This example emphasizes the importance of environment and lifestyle: while genetics contributes to the risk of a disease, it is not deterministic and differences in (or changes to) someone’s environment can increase or decrease their overall risk. Identifying individuals at high genetic risk would allow targeted public health messages and lifestyle interventions to try to prevent disease manifestation.
Any application of PRSs for precision medicine would need to overcome significant challenges. Polygenic medicine represents a paradigm shift for clinicians: most are familiar with rare high-penetrance single gene mutations, but not with genetic predisposition based on a continuous risk score. Education for clinicians and the public will be imperative to hasten clinical uptake. Organizations such as the U.K. Department of Health’s Genomics England, which runs the 100,000 Genomes Project, offer resources to communicate information about genomic medicine with rare variants, but education on polygenic medicine is significantly lacking.
As the predictive accuracies of PRSs improve, commercial interest in them will undoubtedly grow as well. During the past year, the field has seen some interesting developments: in September 2017, Myriad Genetic Laboratories launched riskScore, a product which estimates a woman’s polygenic risk of breast cancer based on 86 genotyped SNPs. A month later, the Scripps Translational Science Institute and The Scripps Research Institute in California released a smartphone app called MyGeneRank, which estimates a person’s polygenic risk of coronary artery
disease based on their 23andMe genetic data. In addition, a senior scientist at Helix, a U.S.-based company that provides direct-to-consumer (DTC) DNA testing as well as a marketplace for products based on DNA tests, was quoted just a few months ago as saying that most DTC DNA testing products will offer polygenic risk scores within the next three years.7
Conclusions
Disease prevention and early disease detection are critical for extending human longevity. During the past few years, hundreds of research papers have been published demonstrating that PRSs can capture important information about an individual’s risk of developing common diseases. For heritable diseases, a person’s genetic risk can be measured from birth (or even before birth); therefore, in theory, a newborn’s PRS could enable disease prediction for him or her long before the typical age of onset of the disease and possibly decades before the disease’s clinical risk factors become apparent.
Many scientific, clinical, and social obstacles must still be overcome to bring PRSs into clinical practice. Further improvements in polygenic risk profiling capabilities are essential. In particular, methods are needed to enable prediction estimates to be adjusted based on an individual’s ancestry, demographics, and clinical information (most genetic research carried out to date has only been among populations of European ancestry). Other improvements might include using whole genome prediction models, machine learning, and artificial intelligence to increase prediction accuracy.
Regardless, the era of polygenic medicine is approaching fast: the leap in the scientific understanding of polygenic risk profiling and the explosion in public interest in genetics have brought us to a point where, in due course, PRSs will almost certainly have a place in clinical risk prediction and potentially in insurance risk stratification, if regulations allow.





